downloading
Downloading sequencing data
https://www.youtube.com/watch?v=fAaVzhD7DOU&ab_channel=WCBbioinformatics
https://docs.google.com/presentation/d/12CJ-8CQSryc3K72kok3BHpw-rxTOv2pN06GJF2FP4xE/edit?usp=sharing
Published data repositories
Published sequencing data is deposited online in
SRA (Sequence Read Archive)
The SRA is a repository for raw sequencing data
- Long and short reads from any platform (Illumina, PacBio..)
Organised with unique identifiers
SRP = Project/Study (1 or more samples )
SRS = Sample (1 or more experiments)
SRX = Experiment (1 or more runs i.e technical replicates)
SRR = Run (results for 1 sequencing run)
Runs stored in sra format
- Reads saved in spots
Smaller datasets and metadata can be downloaded via links in each run
Other download options
SRA Toolkit (see below)
Cloud access
GEO (Gene Expression Omnibus)
Genomics data repository for high throughput experiments
Experimental metadata
Processed data e.g bigWig read profiles, ChIP-seq peak files, RNA-seq read count tables
Links to raw data (SRA)
Links to publication
Data organised into
Series and super series (GSE)
Samples (GSM)
Most publications will contain a GEO series ID.
Download links are available at the bottom of each GEO submission.
To download to the server
Right click to “Copy link as”
Use
wgetorcurlon the command line to download using the URL
ENA (European Nucleotide Archive)
ENA is repository for sequencing data hosted by the European Bioinformatics Institute.
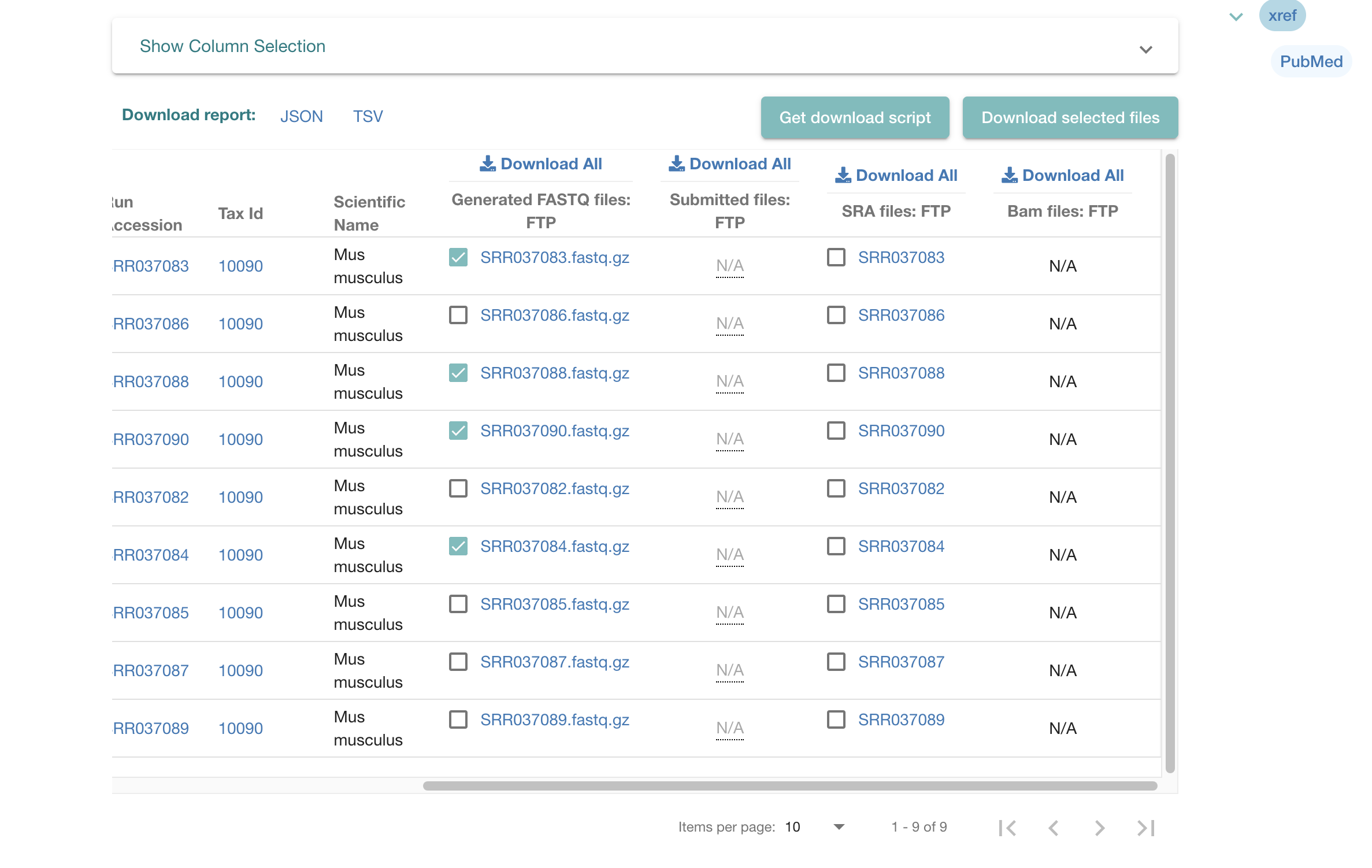
To download raw sequencing files from a project:
Select the fastq files you require.
Hit the “Get Download Script” button.
Run the script on the terminal command line on your computer or on the server.
e.g.
sh ena-file-download-read-run-PRJNA112117-fastq_ftp-20240623-919.sh
SRA Toolkit
https://github.com/ncbi/sra-tools/wiki/08.-prefetch-and-fasterq-dump